Gateway modules
SWEBeams defines in its final report the concept of “Instegsmiljö” – Gateway system, which in the broadest sense describes platforms and constellations that allow the preparation of reagents and/or provide expertise for subsequent experiments in particular at synchrotrons, neutron sources, and NMR, with particular of Swedish focus on MAX IV laboratory and in the future also ESS (Figure 1). PPS features two specializations that cater to produce reagents to use in neutron sources, but also NMR or other types of experiment that uses (per)deuterated proteins in module A. and in B. the production of reagents to use in synchrotrons or Cryo-EM.
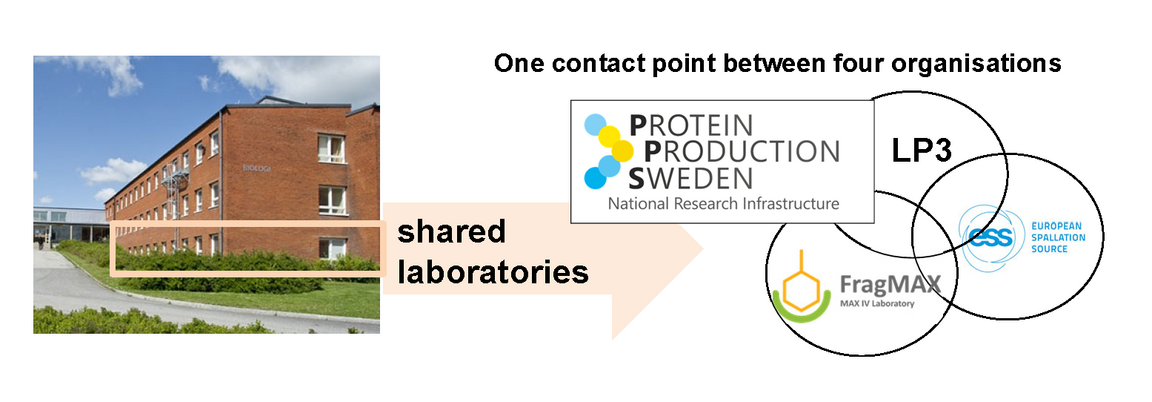
Figure 1.
Biology house A of Lund University is a meeting point of 4 platforms from MAX IV laboratory, the European Spallation Source ERIC and Lund Unviversity, as well as the national research infrastructure PPS.
A) Biological (Per)deuteration – reagents for neutron experiments and NMR
(Per)deuterated proteins and other bio-derived molecules are important for NMR, spectroscopy, neutron reflectometry, small angle neutron scattering, and neutron protein crystallography. In particular, deuterated biological molecules are of utmost importance to neutron scattering experiments. Neutrons and in some cases the scattering magnitude can be quite different for isotopes of the same element. This effect is most pronounced for different isotopes of hydrogen, specifically 1H (protium, H) and 2H (deuterium, D), especially considering the elements found in biological material, where D has higher scattering length and much lower background compared to H. To benefit from these differences, the strategic use of deuterated molecules in neutron scattering experiments add significant information and many experiments are unfeasible without D-labeled materials.
The extent of deuterium incorporation is described as fully deuterated (perdeuterated), partially deuterated (deuterated), or exchange of labile H with D (H/D exchanged). For many types of neutron experiments, it is advantageous to replace as many H with D as possible due to their different neutron scattering properties. However, the exact deuteration grade needed depends on the type of biomolecule, type of experiment and scattering technique used.
For D-labeling of biomolecules there are many strategies, but by far the most commonly used is to culture living cells (e.g. bacteria, yeast, microalgae that can tolerate a deuterated environment) under deuterated conditions. Deuterium oxide (D2O) is toxic to higher organisms, and it is difficult and very expensive to incorporate to >30% uniform D level in living systems.
LP3 at Lund University and now part of PPS has worked with (per)deuteration of proteins for its users since 2013 for in particular for applications in neutron scattering and NMR and collaborates both in the development and implementation of “best practice” for (per)deuteration of biological macromolecules with other laboratories internationally within DeuNet (The Deuteration Network, https://deuteration.org/) and disseminates knowledge about deuteration and neutron scattering experiments [1-5].
What PPS offers:
We offer expertise and reagent production in particular for neutron scattering experiments but also all other experimentation requiring protein labeling, including multiple labeling with 15N and/or 13C. We therefore encourage any requests fitting the broad profile of need of reagents for neutron scattering but also for any other type of experiment in need of (per)deuterated proteins. This includes:
- Production of any protein reagent for neutron scattering experiments
- Feasibility studies for the production of (per)deuterated proteins e.g. using specialized expression system [6] and new technologies [7].
- Production of (per)deuterated proteins for any type of experiments using deuterated proteins (e.g. neutron scattering, MS, spectroscopy NMR (including multiple labeling and labeling with other stable isotopes than D).
B) Molecular Chaperones – reagents for X-ray crystallography or other experiments at synchrotrons or for Cryo-EM
The term molecular chaperones aims at Affinity Probe-Enabled Structural Analysis. In this track of protein production, we aim at to deliver probes, such as antibody fragments, to facilitate structure determination of proteins, the structure of which cannot be determined by conventional methodology. The probes can enable crystallization of proteins (such as membrane proteins) that cannot be crystallized by conventional methodology by acting as crystallization chaperones, thereby enabling downstream structure determination using X-ray crystallography. Such probes can also increase the total mass of proteins by forming a complex, enabling a structural characterization by Cryo-EM.
LP3 at LU collaborates in this PPS module with MAX IV laboratory, the SciLifeLab’s Human Antibody Therapeutics National Facility in Lund and the Lund node of SciLifeLab’s SciLifeLab’s CryoScreeNET.
What PPS offers:
We therefore encourage any requests fitting the broad profile of need of reagents for X-ray crystallography and Cryo-EM using Affinity Probe-Enabled Structural Analysis. This includes:
- Production of any protein complex
- Production of any protein reagent, probe and target protein, for structure determination
- Purification of probe & target protein complex
- X-ray crystallography trials and dataset collection e.g. at MAX IV.
[1] Orozco Rodriguez JM, Wacklin-Knecht HP, Clifton LA, Bogojevic O, Leung A, Fragneto G, et al. New Insights into the Interaction of Class II Dihydroorotate Dehydrogenases with Ubiquinone in Lipid Bilayers as a Function of Lipid Composition. Int J Mol Sci. 2022;23.
[2] Delhom R, Nelson A, Laux V, Haertlein M, Knecht W, Fragneto G, et al. The Antifungal Mechanism of Amphotericin B Elucidated in Ergosterol and Cholesterol-Containing Membranes Using Neutron Reflectometry. Nanomaterials (Basel). 2020;10.
[3] Koruza K, Lafumat B, Nyblom M, Mahon BP, Knecht W, McKenna R, et al. Structural comparison of protiated, H/D-exchanged and deuterated human carbonic anhydrase IX. Acta Crystallogr D Struct Biol. 2019;75:895-903.
[4] Koruza K, Mahon BP, Blakeley MP, Ostermann A, Schrader TE, McKenna R, et al. Using neutron crystallography to elucidate the basis of selective inhibition of carbonic anhydrase by saccharin and a derivative. J Struct Biol. 2019;205:147-54.
[5] Koruza K, Lafumat B, Végvári Á, Knecht W, Fisher SZ. Deuteration of human carbonic anhydrase for neutron crystallography: Cell culture media, protein thermostability, and crystallization behavior. Arch Biochem Biophys. 2018;645:26-33.
[6] Kelpšas V, Wachenfeldt CV. Strain improvement of Escherichia coli K-12 for recombinant production of deuterated proteins. Sci Rep. 2019;9:17694.
[7] Koruza K, Krupinska E, Sele C, Végvári Á, Knecht W, Fisher SZ. Botryococcus braunii autolysate for the production of deuterium-labeled recombinant protein. Algal Research. 2024;79.